UPDATE: After playing around with MobileNet and DenseNet, I've come to the conclusion that DenseNet had better and more robust performance in terms of log-loss, AUC(90% val/92% test), and F1 score(86% val/88% test). I updated my github repo with the codes for training a 3-class classification model. The final metrics were calculated only based on the melanoma class as if it's a binary classifier.
The largest public collection of dermoscopic images of skin lesions is maintained by International Skin Imaging Collaboration (ISIC). The images were collected from established clinical centers worldwide, and they were captured by various devices within each center. The goal of ISIC's collaboration effort can be summed up in this statement:
"As inexpensive consumer dermatoscope attachments for smart phones are beginning to reach the market, the opportunity for automated dermoscopic assessment algorithms to positively influence patient care increases."
MobileNet - one channel at a time
With that in mind, I want to train a lightweight deep neural net called MobileNet with dermascopic images, and see how it performs as a binary classifier (see complete training code). What makes MobileNet special is its small and elegant architecture optimized for mobile devices, while not compromising too much in accuracy as shown in its ImageNet performance . Instead of using regular convolution layer to learn image features(i.e., shape, area), it uses depthwise convolution followed by point-wise convolution to dramatically reduce computational cost (Fig.2). MobileNet is loaded with 4.3M parameters, compare to InceptionV3 which has 23.9M parameters.
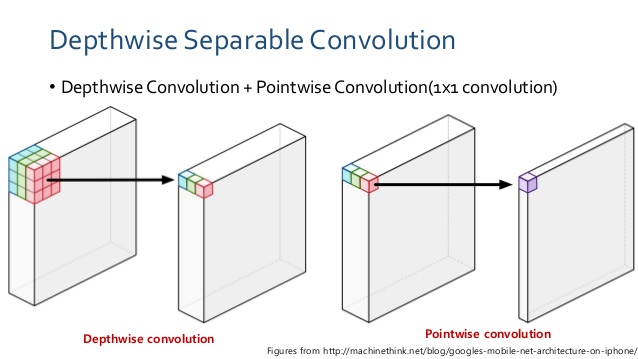
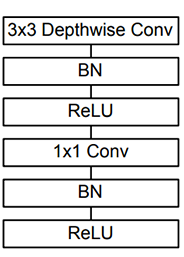
A regular convolution both filters and combines inputs into a feature map in one step. In contrast, depthwise separable convolution performs convolution on each channel separately first, effectively filters and selects features for the channel, then applies pointwise (1x1) convolution to merge the features to generate a new feature map. According to the Mobilenet paper [2], for a 3×3 kernel depthwise separable convolution is about 9X faster than regular convolution operation. MobileNet has 28 layers, most of which are repeats of depthwise separable convolution, and 95% of the total computation is spent on the 1x1 pointwise convolution[2].
Data Processing
The dermoscopic images had many distracting artifacts (e.g, vignette borders, measurement scale, hair, etc) that can possilby interfere with the model learning. Instead of writing a code for each type of artifacts to clean up the image, I realize it's more efficient to apply binary segmentation mask to select for the general lesion area. For segmentation, I used the SegNet autoencoder model where it achieved a Dice coefficient (metric of overlap) of 78% on validation data.
Another issue is that the malignant category was underpresented (20%), and it's difficult to train a deep neural net model with fewer than 1,000 pictures per class(yes, I did try). I used augmentation (rotation and translation) to produce copies of the original malignant images to mitigate the under-representation. No other augmentation methods were used at this stage.
Transfer the learning with Bottleneck
There's a wonderful tutorial by the creator of Keras illustrating transfer learning for deep neural net. What it means is that you take advantage of fine-tuned model trained on large datasets, and start training new data with the pre-trained weights to save time and computational cost. One common way to do that is using 'bottleneck' features - output of the 'bottom' part of a deep learning model, essentially it's all the convolutional layers (everything up to the fully-connected layers ). I saved the bottleneck features as numpy array, then train a customized 'top' part of the model for a binary classifier (see code below). Training the top part separately ensures that the top model weight provides a consistent and smooth training when it's attached to the bottom model later on.
After training the top model, I augmented the image files with ImageDataGenerator() along with
the default preprocessing function to scale the pixel values. I chose Stochastic Gradient Descend as the optimizer
because that was the optimizer used on the original MobileNet.
Binary cross entropy was a loss function used to optimize the model(the lower the score the better), it takes into account confidence of the prediction along with accuracy. The validation binary cross entropy was 0.43, F1 score (balanced accuracy accounting for imbalanced class distribution) was 86%, and AUC for ROC was 0.89 (1 being the best).
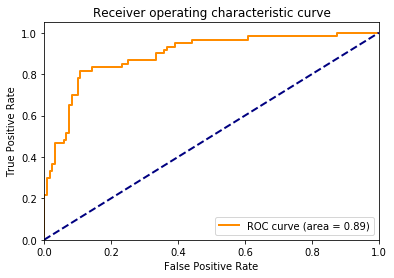
While the AUC score was lower than the ones presented by Esteve et al[4], I should point out that this MobileNet model was trained on a smaller training dataset provided by ISIC 2017 Challenge. I expect that better performance (>90% balanced accuracy) can be achieved with a larger training set, as well as bigger image array. For my own future reference, here are some things to keep in mind when training a deep learning model for image classification:
- Pay attention to preprocessing step for a given deep neural net model. It's NOT always mean centered or standard deviation normalized to 1.
- Sanity check: make sure the images in trainnig and test sets have more or less the same distribution.
- Explore augmentation options such as contrasting or adaptive histogram equilization. It is a handy tool especially when poor lighting or resolution of the digital image becomes an issue (Note to self: need to explore this further).
- Obviously there's room for improvement. For example,the number of filters in each layer and dropout rate can be tuned.
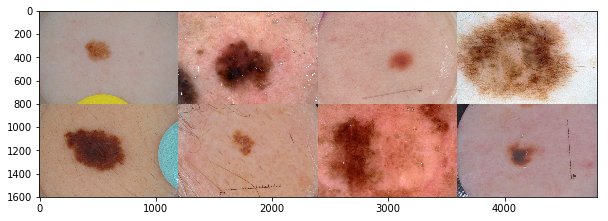
Note: For Fig.1, the center 4 images are melanoma, and the rest are benign.
REFERENCES
[1]Herman, C. Emerging Technologies for the Detection of Melanoma: Achieving Better Outcomes. Clinical, Cosmetic and Investigational Dermatology 5 (2012): 195–212.
[2]MobileNets: Efficient Convolutional Neural Networks for Mobile Vision Applications(https://arxiv.org/pdf/1704.04861.pdf)
[3]Keras Documentation: https://keras.io/applications/
[4]Esteva, A.,Kuprel, B., et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 542(2017): 115–118.
